miRnomics Atlas
Genetic regulation of microRNAs
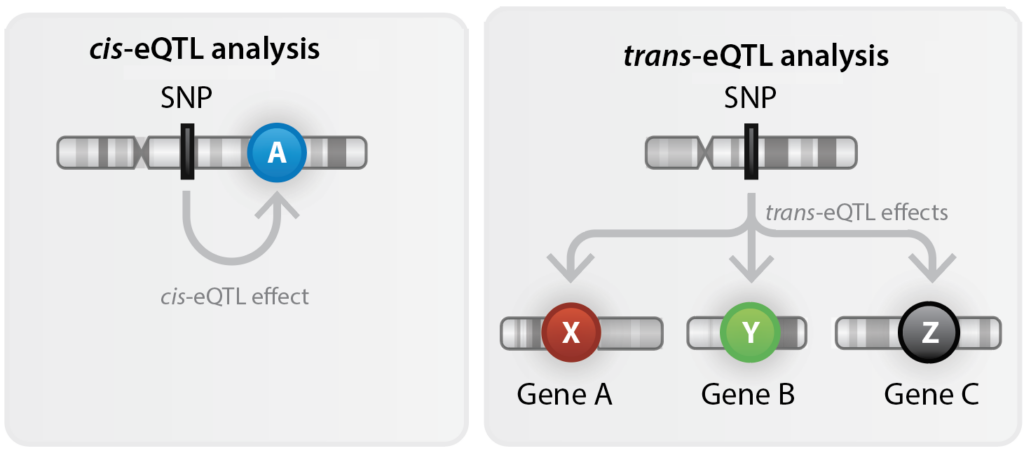
Genetic variants in our genome could influence the transcription or function of miRNAs, affecting miRNA-mediated gene regulation and leading to diseases. Integration of genomics and miRNA expression data enables interrogation of the genetic architecture of miRNAs, and provides valuable resources for future studies on clinical significance of miRNAs in human disease. Here, we present the results of our genome-wide association studies on over 2000 extracellular miRNAs circulating in plasma measured by RNA-sequencing in the Rotterdam Study (Phase 1). We report both cis and trans miRNA-expression quantitative trait loci (miRNA-eQTLs) at the nominal significance level (p-value < 0.05) and those passing the significance threshold of 1e-5, also the SNPs that have been replicated across two independent cohorts (see Ref below). The consequences of perturbation in miRNA transcription on a wide range of clinical conditions were also systematically investigated in phenome-wide association studies (PheWAS), with their causality tested using Mendelian randomization (MR) in the UK Biobank. Moreover, our colocalisation studies have highlighted several miRNA-eQTLs overlapping with gene expression, protein, and metabolite-QTLs and with disease-associated variants reported in previous GWAS studies (see the disease association and omics integration parts).
SNP
Search all data using a SNP ID
Locus
Search all data using a chromosome position
Cis-eQTL
Search for 1000 top cis-miRNA-eQTLs
DataBase
Download
Download Description
You are able to access the summary statistics of genome-wide association studies (GWAS) conducted on plasma levels of 2083 miRNAs in the Rotterdam Study cohort, which consists of ~2,200 participants through the link below (for SNPs with p-value < 1e-5). The summary stat data including SNPs associated with miRNAs at a nominal p-value (< 0.05), after filtering out SNPs with an Rsq value higher than 0.7 and a minor allele frequency (MAF) greater than 0.01 are available through Zenodo open repository https://zenodo.org/record/13869398. You can download all miRNA-eQTLs (including cis-miR-eQTLs and trans-miR-eQTLs) at once.
Citation
If you use the data on this website, please cite our paper: Mustafa R., (…), Ghanbari M. A comprehensive study of genetic regulation and disease associations of plasma circulatory microRNAs using population-level data. Genome Biology 2024 Oct 21;25(1):276. doi: 10.1186/s13059-024-03420-6.
Contact
For questions, bug reports, or requests, please contact Dr. Mohsen Ghanbari (m.ghanbari@erasmusmc.nl).
Follow US
on These Social Media or Websites