About miRNomics
The miRNomics atlas is a publicly accessible web-based tool developed by Dr. Ghanbari’s group, at the department of Epidemiology, Erasmus MC Rotterdam, the Netherlands. Its purpose is to disseminate the findings of their pioneering research on microRNAs using large-scale population-level data. The research encompasses various aspects, including the genetic regulation of miRNAs, the association of miRNAs with diverse clinical outcomes, and the integration of miRNAs with other omics (incl. genes, proteins, metabolites) to identify miRNA-regulated pathways. This atlas aims to elucidate the role of miRNAs in the pathogenesis of various complex diseases and explore their potential as biomarkers.
The research conducted by Dr. Ghanbari’s group utilizes population-based omics data and employs advanced analytical approaches, which is followed by in-vitro validation studies for the best candidate miRNAs. The results of their projects in the emerging field of miRNA-gene regulation are categorized into three parts: A) Genetic regulation, B) Disease association, and C) Omics integration of miRNAs. To access their projects and publicly available data, you can click on the provided links within the miRNomics atlas.
miRNomics
Our Studies
Disease association of miRNAs
Omics integration of miRNAs
microRNAs
Biogenesis and Function
microRNAs (miRNAs) are small, evolutionary conserved, single-stranded, non-coding RNA molecules that range between 18-24 nucleotides in length. They play a crucial role in post-transcriptional gene expression regulation. The discovery of miRNAs dates back to the early 1990s when they were first identified in the organism C. elegans. To date, more than 2500 mature miRNAs have been identified in the human genome, with the majority of them exhibiting high conservation across other animal species. Collectively, miRNAs have the potential to regulate the expression of over half of the coding genes in humans. Read more about miRNAs and the Nobel Prize in Physiology or Medicine 2024 for the discovery of miRNAs in the following link: The Nobel Prize in Physiology or Medicine 2024 – NobelPrize.org
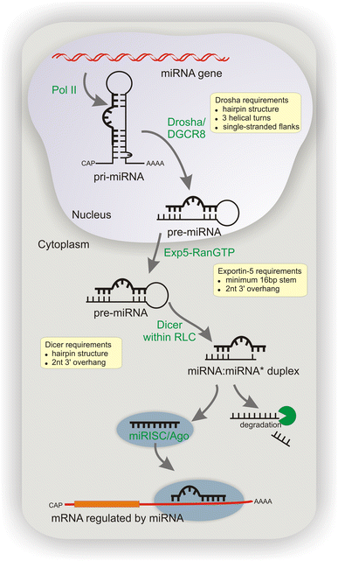
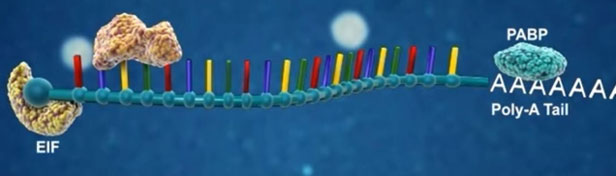
Canonical miRNA biogenesis begins with the transcription of DNA sequences into primary miRNAs (pri-miRNA). The microprocessor complex, composed of Drosha and DiGeorge Syndrome Critical Region 8 (DGCR8), cleaves the pri-miRNA in the nucleus, generating the precursor-miRNA (pre-miRNA). The pre-miRNA is then exported to the cytoplasm in an Exportin5/RanGTP-dependent manner and processed by DICER, resulting in the production of the mature miRNA duplex. Subsequently, either the 5p or 3p strand of the mature miRNA duplex is incorporated into the Argonaute (AGO) family of proteins, forming a miRNA-induced silencing complex (miRISC). In most cases, the miRISC binds to target messenger RNAs (mRNAs) through base-pairing interactions between the miRNA and complementary sequences in the 3′ untranslated region (UTR) of the mRNAs. This interaction leads to gene silencing through mRNA cleavage, destabilization through shortening of the polyA tail, or inhibition of mRNA translation into proteins. Nevertheless, interactions between miRNAs and other regions, including the 5′ UTR, coding sequence, and gene promoters, have also been reported. The interaction between miRNAs and their target genes is dynamic and influenced by various factors, such as the subcellular localization of miRNAs, the abundance of miRNAs and target mRNAs, and the affinity of miRNA-mRNA interactions.
miRNAs can be released into extracellular fluids (e.g., plasma, cerebrospinal fluid, saliva, breast milk, urine) and transported to target cells through vesicles, such as exosomes, or by binding to proteins, including Argonauts. These extracellular miRNAs act as chemical messengers, facilitating cell-to-cell communication. Notably, extracellular miRNAs, particularly those circulating in serum or plasma, exhibit high stability and hold potential as biomarkers for various diseases.
External Page Click here
Association of miRNAs with
Clinical traits and Diseases

- The association of miRNAs with complex traits and diseases can be investigated through various approaches. In our studies, we have explored the association and potential role of numerous miRNAs with a wide range of clinical outcomes. We achieved this by utilizing omics data from large-scale population-based cohort studies, as well as publicly available genetic data. The findings of our studies are compiled in this section, and you can search for specific miRNAs or traits/diseases by entering their names.
HOW IT WORKS
Join us with three
easy steps
Quantum porter
Lorem ipsum dolor sit amet, consectetur adipiscing
elit. Ut elit tellus, luctus nec ullamcorper mattis,
pulvinar dapibus leo.
Quantum porter
Lorem ipsum dolor sit amet, consectetur adipiscing
elit. Ut elit tellus, luctus nec ullamcorper mattis,
pulvinar dapibus leo.
Quantum porter
Lorem ipsum dolor sit amet, consectetur adipiscing
elit. Ut elit tellus, luctus nec ullamcorper mattis,
pulvinar dapibus leo.
Our Customers
Testimonials


Postdoctoral researcher, Department of Epidemiology, Erasmus MC University Medical Center, Rotterdam, the Netherlands
Mohen Ghanbari
July 2017- May 2019
Assistant Professor and PI of Molecular and Systems Epidemiology, Department of Epidemiology, Erasmus MC, Rotterdam, the Netherlands
Mohen Ghanbari
Dopel limited
Associate Professor and PI of Molecular and Systems Epidemiology, Department of Epidemiology, Erasmus MC, Rotterdam, the Netherlands
Mohen Ghanbari
June 2019 – October 2022

Follow US
on These Social Media or Websites